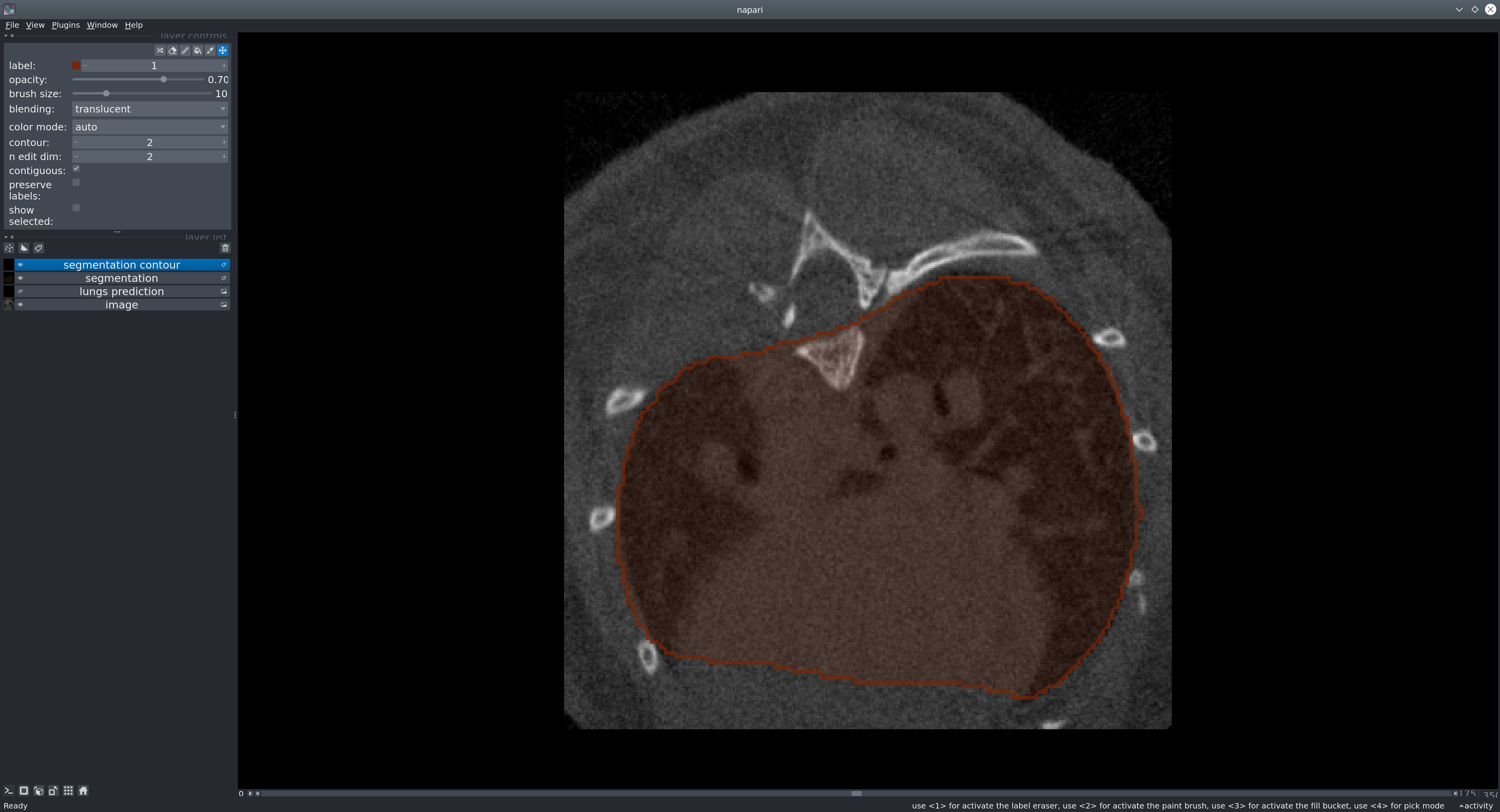
Neural network for segmenting mouse lungs in CT scans, based on the classic U-Net architecture.
The goal is to provide a reliable, easy-to-use tool to obtain binary lung masks from mouse CT scans.
- Try in your browser (Hugging Face Space)
- Installation
- Use in napari (GUI)
- Use as a Python library
- Use from the command line
- Run with Docker (GHCR)
- Models & weights
- Dataset
- Issues
- License
- Acknowledgments
- Carbon footprint
No local installation needed — test the model directly in your browser:
➡️ https://huggingface.co/spaces/qchapp/3d-lungs-segmentation
Upload a mouse CT scan, run the segmentation, visualize and download the resulting lung mask.
We recommend using a fresh Python environment.
- Requirements:
python>=3.9,pytorch>=2.0
Please install PyTorch first for your platform following the instructions on pytorch.org.
Install the package from PyPI:
pip install unet_lungs_segmentationOr install from source:
git clone https://github.com/qchapp/lungs-segmentation.git
cd lungs-segmentation
pip install -e .Napari is a multidimensional image viewer for Python.
-
Launch napari:
napari
-
Open an image via File → Open files… or drag & drop into the viewer.
To open formats like NIfTI directly, consider installing the pluginnapari-medical-image-formats. -
Sample data: In napari, go to File → Open Sample → Mouse lung CT scan to try the model.
-
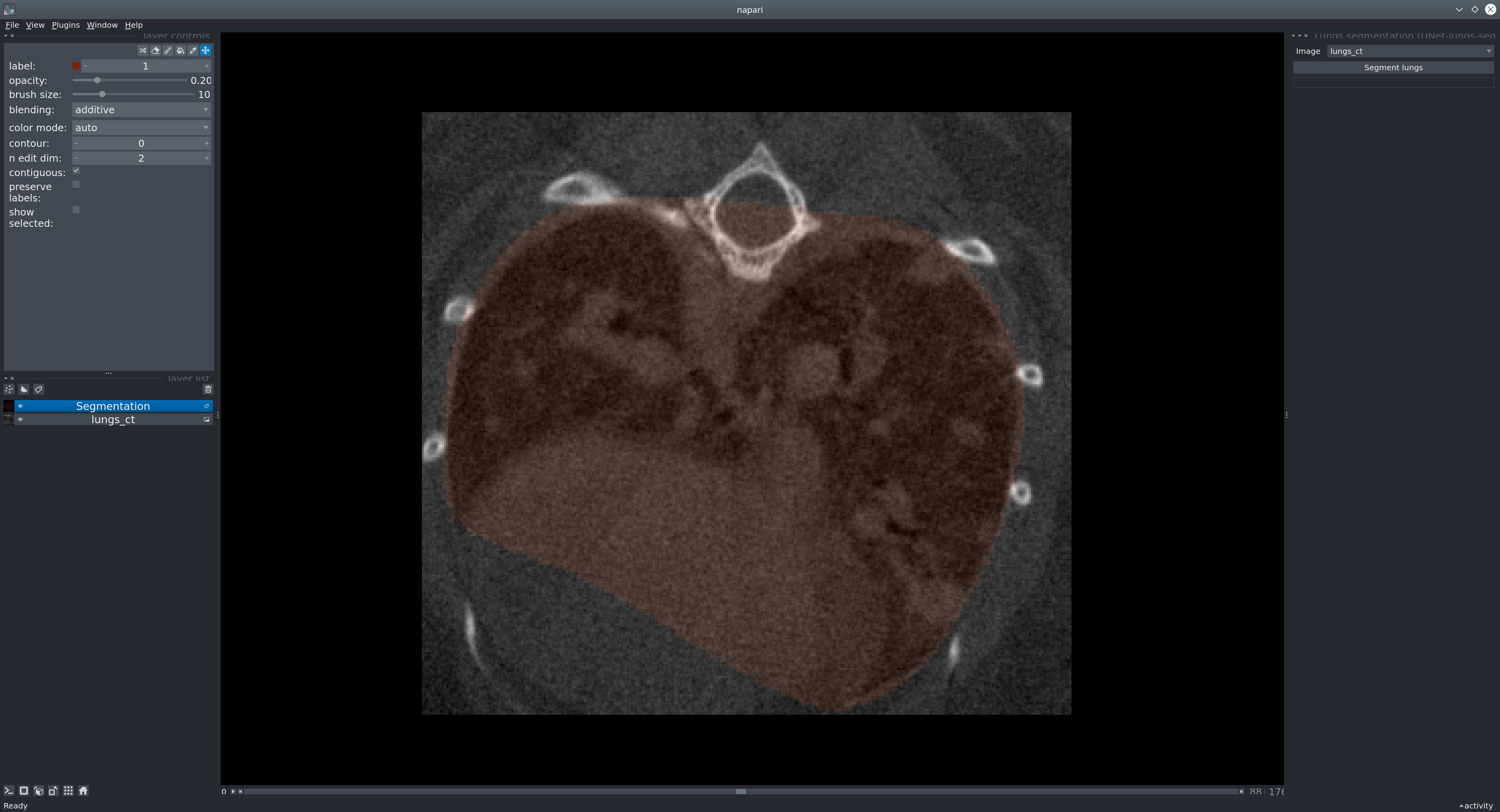
Run the plugin via Plugins → Lungs segmentation (unet_lungs_segmentation).
Select the image layer and click Segment lungs.
Run the model in a few lines to obtain a binary mask (NumPy array):
from unet_lungs_segmentation import LungsPredict
lungs_predict = LungsPredict()
mask = lungs_predict.segment_lungs(your_image) # your_image: NumPy arraySpecify a custom probability threshold (float in [0, 1]) if desired:
mask = lungs_predict.segment_lungs(your_image, threshold=0.5)Run inference on a single image:
uls_predict_image -i /path/to/folder/image_001.tif [-t <threshold>]- If
-t/--thresholdis provided, it binarizes the prediction at that value (default:0.5in[0,1]). - The output mask is saved next to the input image:
folder/
├── image_001.tif
└── image_001_mask.tif
Batch-process a folder:
uls_predict_folder -i /path/to/folder/ [-t <threshold>]Produces, e.g.:
folder/
├── image_001.tif
├── image_001_mask.tif
├── image_002.tif
└── image_002_mask.tif
You can run the tool without a local Python setup using our container on GitHub Container Registry.
- Pull the image:
docker pull ghcr.io/qchapp/unet-lungs-segmentation:latest- Run on a single image (mount a data folder into the container):
docker run --rm -v /path/to/data:/data ghcr.io/qchapp/unet-lungs-segmentation:latest \
-i /data/image_001.tif -t 0.5- Run on a folder:
docker run --rm -v /path/to/data:/data ghcr.io/qchapp/unet-lungs-segmentation:latest \
uls_predict_folder -i /dataNote: The container’s entrypoint is
uls_predict_image.
For folder mode, prefix the command explicitly as shown above.
The model weights (~1 GB) are downloaded automatically on first use from Hugging Face.
The model was trained on 355 images from 17 different experiments and 2 scanners, and validated on 62 images.
If you encounter problems, please open an issue with a clear description and, if possible, a minimal reproducible example.
This project was developed as part of a Bachelor’s project at the EPFL Center for Imaging.
It was carried out under the supervision of Mallory Wittwer and Edward Andò, whom we sincerely thank for their guidance and support.
This model is released under the BSD-3 license.
As estimated by Green Algorithms, training this model emitted approximately 584 g CO₂e.