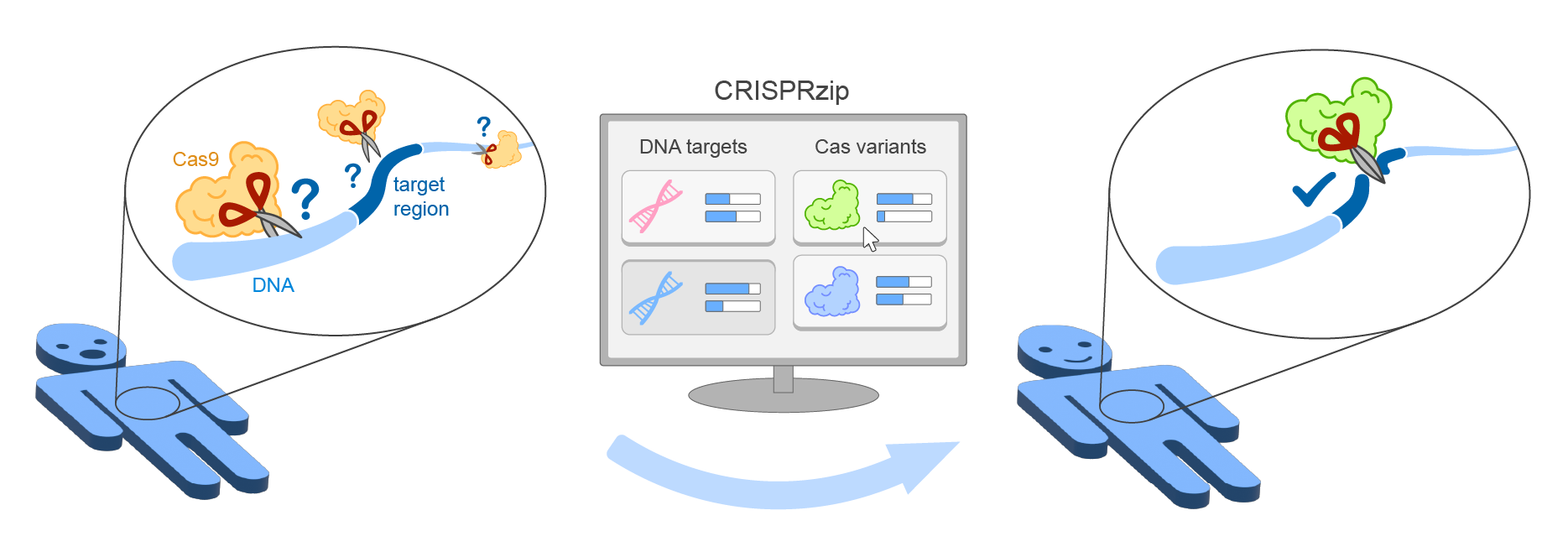
Welcome to the codebase of CRISPRzip from the Depken Lab at TU Delft.
CRISPRzip is a physics-based model to study the target recognition dynamics of CRISPR-associated nucleases like Cas9 (Eslami-Mossalam, 2022). Their interactions with target DNA is represented as an energy landscape, with which you can simulate binding and cleavage kinetics. The parameters have been obtained by machine learning on high-throughput data. CRISPRzip makes quantitative predictions of on-target efficiency and off-target risks of different guide RNAs.
With CRISPRzip, we hope to contribute to assessing the risks that come with particular choices in CRISPR application, and as such contribute to the development of safe gene editing technology.
Eslami-Mossallam B et al. (2022) A kinetic model predicts SpCas9 activity, improves off-target classification, and reveals the physical basis of targeting fidelity. Nature Communications. 10.1038/s41467-022-28994-2
CRISPRzip is available on PyPi and can be installed with pip.
Although CRISPRzip can be directly installed from pip, creating a virtual environment makes it easier to manage dependencies. Below you can see some intructions on how to generate a virtual environment with venv assuming you already have python installed in a bash-like terminal.
python -m venv crisprzip-venv
source crisprzip-venv/bin/activate
pip install crisprzipTo be able to make changes and contributions to CRISPRzip, you will need to get your own copy of the source code and install software dependencies on your own. Assuming you have a python and git installed in a bash-like terminal, the installation process can be done with the following instructions.
git clone https://github.com/hiddeoff/crisprzip.git
cd crisprzip
python -m venv crisprzip-venv
source crisprzip-venv/bin/activate
pip install -e .Please use our contributing guidelines if you would like us to consider your developments in a future CRISPRzip release
CRISPRzip development includes a cross-platform compatibility and test workflow. If you would like to verify your local installation, please follow the Developer Installation instructions, then run the following test.
source crisprzip-venv/bin/activate
pip install -e '.[tests]' # installs pytest and pandas
pytest tests/cleavage_binding_prediction/test_cleavage_binding_prediction.py -vYou can also follow the user installation and execute the code in the Usage section below.
CRISPRzip makes predictions about cleavage and binding activity on on- and off-targets. First, you define the protospacer and target sequence, and then, you can predict the fraction cleaved or bound.
# 1. load parameter set
from crisprzip.kinetics import load_landscape
searcher = load_landscape("sequence_params")
# 2. define Cas9, gRNA and DNA target
searchertargetcomplex = searcher.probe_sequence(
protospacer = "AGACGCATAAAGATGAGACGCTGG",
target_seq = "AGACCCATTAAGATGAGACGCGGG", # A13T G17C
)
# 3. predict activity
f_clv = searchertargetcomplex.get_cleaved_fraction(
time=600, # 10 minutes
on_rate=1E-1
)
f_bnd = searchertargetcomplex.get_bound_fraction(
time=600, # 10 minutes
on_rate=1E-1
)
# 4. format output
print(f"After 10 minutes, the target (A13T G17C) is ...")
print(f"- cleaved for {100 * f_clv:.1f}% by Cas9")
print(f" or ")
print(f"- bound for {100 * f_bnd:.1f}% by dCas9")Output:
After 10 minutes, the target (A13T G17C) is ...
- cleaved for 10.5% by Cas9
or
- bound for 94.2% by dCas9
See the tutorial or the docs for more examples how to explore sequence, time and concentration dependency.
We encourage contributions in any form - reporting bugs, suggesting features, drafting code changes. Read our Contributing guidelines and our Code of Conduct.
Many thanks to Elviss Dvinskis, Raúl Ortiz and Aysun Urhan from the DCC team at TU Delft for their support to get this package released!
Technische Universiteit Delft hereby disclaims all copyright interest in the program “CRISPRzip” (a physics-based CRISPR activity predictor) written by the Author(s). Paulien Herder, Dean of Applied Sciences
(c) 2024, Hidde Offerhaus, Delft, The Netherlands.